1st Place Essay
How can DNA help us to discover ancient human history?
Sheen Gahlaut, Claire Surgeoner
The Perse School, Cambridge, United Kingdom
Analysing both modern genomes and ancient DNA (aDNA) extracted from fossils has revolutionized the study of ancient human history by revealing novel phylogenetic relationships, illustrating migration patterns 1 and providing new insights into complex trait evolution 2 . Ancient DNA analysis is rapidly advancing: whilst initially based only on mitochondrial DNA, reliable sequencing of entire genomes is now possible through advances in bioinformatic analysis of degraded DNA molecules, as well as better contamination prevention 3,4,5 . A key advantage of using aDNA is its ability to provide a temporal scale that modern DNA cannot, thus allowing the exploration of a human demographic history which is located in time. Many significant discoveries involve archaic humans preceding Homo sapiens . aDNA has surprisingly identified that admixture did indeed occur between humans and Neanderthals, with 4% of modern human DNA outside Africa being of Neanderthal origin – geographic patterns of genetic variation suggest this was a result of humans migrating out of Africa meeting with Neanderthals in the Middle East, then further migration across Asia and Europe 6 . Admixture also occurred between humans and Denisovans, another archaic human group that was discovered using aDNA: as very few physical remains exist, only genomic data highlights their contribution to the ancestral populations in Southeast Asia 7 . Both these and many other discoveries were made possible by comparing modern human genomes and ancient genomes, illustrating the importance of utilising both in combination. Admixture studies also illustrate how ancient humans were able to survive and colonise vast areas: studies of aDNA highlight the importance of historical interbreeding in providing humans with critical disease-fighting genes 8 .
Ancient DNA is also able to provide insights into ancient human migration, and can augment understanding of change as a result of natural selection based on the geographical location of different groups. Overall, it is evident that there was a much greater amount of large-scale migration (and mixing of different populations) than previously considered to be the case. Additionally, aside from allowing us to discover broad ideas about ancient human history, DNA can also unravel the histories of individual groups of people and the spread and subsequent development of their cultures. The steppes of Central Asia (previously populated by hunter-gatherer people) became home to the Yamnaya 5,300 years ago, who had a culture characterised by expansion using new technology such as carts. Using only archaeological data, this pattern could result from either the movement of ideas/technologies or of people. Only aDNA was able to confirm that this was due to the simultaneous migration of the Yamnaya people all across Europe and Asia. 9 Findings such as these are not only significant in understanding how ancient human populations migrated, but also in how those movements led to both cultural changes (e.g. the growth of Indo-European languages can be attributed to the migration of the Yamnaya people 7 ) and social developments – e.g. migration patterns of each sex can be considered to better understand the development and persistence of social roles and inequality. 10
Finally, aDNA plays a key role in our understanding of the evolution of complex traits (traits influenced by many genes and environmental factors 11 ) such as height and pigmentation of hair/skin. Comparison of ancient and modern genomes can even illustrate the independent evolution of certain traits in humans and Neanderthals. For example, a fragment of Neanderthal DNA contained a version of the mc1r gene with a changed base-pair which is present only at a very low frequency in humans (evidenced by comparison of that fragment with 3,700 modern human mc1r sequences) 12 . Such individual phenotypic developments can illustrate the importance of various characteristics and the effects of natural selection in ancient human history. Further examples also illustrate more abstract concepts. The FOXP2 gene, present in different forms in humans and chimps, is also present in Neanderthals, and is more similar to the newer human version of the gene, as opposed to the more probable similarity to the older chimp gene 13 ; whilst this is not conclusive in identifying the linguistic ability of Neanderthals, discoveries like this can provoke new ideas and subsequently new questions in research.
Overall, it is evident that DNA can provide us with an extensive range of new discoveries about ancient human history through analysis and comparison of ancient and modern DNA; such discoveries can range from analysis of phenotypes and natural selection to migration, admixture, and the development of culture.
2nd Place Essay
How can DNA help us to discover ancient human history?
Sneha Goni, Nicole Copeland
The Perse School, Cambridge, United Kingdom
DNA sequencing has a wide range of applications from diagnosing diseases to revealing events from the past. Recently, the scale of DNA sequencing has grown rapidly, both in the sequencing of modern human DNA as well as ancient DNA (aDNA), offering a multitude of opportunities. New methods, such as next-generation sequencing, allow researchers to overcome the challenge of DNA contamination in order to sequence the whole genome of aDNA, providing greater insight into ancient human history [1]. Researchers have also discovered that we can extract large amounts of aDNA from the petrous [2]. Since sections of DNA are passed through generations, scientists can determine how closely related individuals are by comparing their DNA sequences; more similar DNA sequences mean individuals are more closely related and share a more recent common ancestor [3]. Firstly, analysing DNA allows us to determine the migration patterns of our ancestors. For example, studies reveal a lower genetic diversity in populations outside of Africa, implying that some early humans migrated out of Africa and settled in other parts of the world. Genetic diversity can be measured by “counting heterozygotes for all variants” [4, p.1]. Moreover, genetic studies also reveal the mixing of early humans with other populations. For example, evidence suggests that around 1.5 to 2.1% of the genome of non-Africans is similar to that of Neanderthals [1], indicating that these individuals lived in similar locations in the past and mixed. Sections of inherited DNA become smaller in each subsequent generation [3] and thus the size of various sections can be used as a temporal measure to establish how long ago an admixture event occurred [2]. On a similar note, DNA analysis can reveal when certain groups diverged from a population and began to settle in other areas. For example, studies comparing genomes demonstrate that a group called the Denisovans split from the Neanderthals later than our distant early human ancestors since their genome is more similar to Neanderthals [2]. Often family trees are used to display the evolutionary relationships between species and populations, showing common ancestors and points at which groups diverged. It is likely that as research continues, new DNA evidence will be discovered, potentially allowing us to identify new species that are currently unknown. Furthermore, studies of DNA allow us to estimate the population size at various times in the past using the ‘Skyline-Plot Method’ which is based on the idea that in smaller populations it is more likely that “two gene copies coalesce to the same ancestral gene copy” [6, p.7]. This may reveal challenges or selective pressures that our ancestors faced, or perhaps implies that certain groups became isolated communities due to geographical barriers, offering an explanation for the changes in population size. However, critics argue that we cannot draw definitive conclusions about population demographics from the limited aDNA data currently available [5]. Studies of environmental DNA can show patterns of land use in the past [6], indicating the ways of life at different times. Another important aspect of ancient human history is the effect and spread of various diseases worldwide and it has now been proposed that DNA studies can offer insight into this [7]. One example of this is the positive selection of genes relating to immunity in some European genes caused by the Black Death, which remains observable in our DNA [8]. Moreover, by sequencing the genomes of pathogens from ancient remains, researchers can understand the spread and influence of disease on ancient humans. Finally, aDNA sequencing can reveal the natural selection of certain genetic variants helping “populations to adapt to local environments” [9, p.6]. For example, the introduction of lighter skin colour in Europeans is thought to be an adaptation to lower sunlight levels, aiding Vitamin D production [8]. Similarly, the increase in the mutation enabling lactose tolerance, resulting from the LCT gene, may be linked to the Neolithic transition (a shift to domestication and farming) [8]. Consequently, genomic studies allow us to infer the types of conditions that ancient humans were living in, by analysing the genetic variants contributing to the observed adaptations. To conclude, DNA analysis allows us to obtain a great deal of information about the lives of ancient humans and population demographics. By piecing together data from various studies and DNA analyses, researchers can understand how people migrated over time, identify instances where they interacted and mixed and uncover information about their environment. In the future, with further technological advances, perhaps we will be able to obtain even more information from DNA.
3rd Place Essay
How can DNA help us to discover ancient human history?
Alicia Reeves-Toy, Astrid Edmunds
Wellington College, Crowthorne, United Kingdom
How can DNA help us to discover ancient human history?Where previously ancient hominids were identified by morphology and location of discovery, DNA analysis has already proved to have unique ability to identify links and differences between species that would otherwise go unnoticed, especially in smaller or fragmented samples. Without DNA analysis we would not know about the existence of Denisovans [1] or that all modern humans share one common ancestor known as ‘Mitochondrial Eve’[2], displaying the impact that DNA analysis can have on our understanding of Ancient Human evolution. DNA analysis is very versatile. Almost all methods of DNA sequencing involve a method of gene amplification which allows even the smallest samples to be identifiable in a laboratory by reproducing the targeted genes [3]. These genes are usually amplified via PCR (polymerase chain reaction).There is, however, some benefit in the use of paleo-proteomics compared to DNA as proteins degrade less readily than DNA itself. For example, a Denisovan jawbone found in a Tibetan cave in 2019 did not have salvageable DNA but could be identified by the proteins found deep within a tooth [5]. While occasionally more useful, this method has fundamental limitations. Crucially, in this discovery, there were only around 2000 amino acids present in the Denisovan jawbone where just one amino acid differed from Neanderthal and modern human sequences. Taken alone, this information would imply the species were far more similar than other methods have proven. Conversely, ancient genomes have contained around 3 million variants compared to any other genome, showing far more detailed variation and heritage. Proteomics and morphology are also both limited in their ability to represent the non-phenotypic information contained within the organism’s cells. Over 98% of the human genome is non-coding DNA, and we are only now beginning to investigate its contributions. Although its composition would not change the proteins formed or any phenotypic features as it doesn’t code for a functional protein, this DNA holds information that could be key to filling in the gaps in our evolutionary picture that morphology, proteomics, and even more traditional DNA analysis (focused only on gene expression) would struggle to identify. Some geneticists believe these ‘proto-genes’ could be raw material used in the synthesis of new genes. This is similar to the ORFs (open reading frames) studied in yeast where there are hundreds of thousands of sequences that could theoretically code for a protein but have no clear function in the context of closely related organisms. Despite this, some strands were still translated, suggesting an alternative purpose [6]. Creating genes from these non-coding regions has the potential to be a strong driving force in evolution as synthesising new genes from this ‘reservoir’ of proto-genes would produce more markedly different proteins than the conservative gene duplication method previously credited as the sole mechanism for evolution. Named de novo genes, these have been identified in several organisms so far including the Atlantic codfish: this has evolved to have an anti-freeze protein that binds to ice crystals in its blood but is not found in any of its ancestors or close relatives. Although quantifying and verifying which genes are de novo is very difficult with current organisms and even harder in ancient samples, technology in the field of genetics is always rapidly developing and will likely soon provide the accuracy and speed needed. De novo genes are a new concept and still largely a mystery however their discovery suggests that genes can be produced from scratch. This has the potential to uncover the key moments of genetic divergence in human history: it could be speculated that even our first separation from the apes could be attributed to de novo genes. When discovering Ancient Human History, DNA analysis has largely succeeded where other methods have failed and will continue to expose a more accurate picture of the evolution of homo sapiens.
3rd Place Essay
How can DNA help us to discover ancient human history?
Ignas Karvelis
Kaunas Jesuit high school, Kaunas, Lithuania
Approximately 200 000 years ago, the first Homo Sapiens evolved in the East Africa region. Since then, we, humans, have spread throughout the world and become the dominant species, however, even though we live in technologically advanced periods our knowledge of the was fairly limited. The first 98 percent of human history is virtually unrecorded. Therefore, scientists require other methods to study ancient human history. This is where DNA, or deoxyribonucleic acid, becomes very important. DNA is a molecule composed of two polynucleotide chains that coil around each other and form a double helix structure. For decades, DNA analysis has allowed for major breakthroughs in the branches of genetics and anthropology. But how does DNA reveal our ancestors’ past?
Part of the answer is in DNA’s purpose. Its function is to carry the genetic information crucial for all living organisms to develop, function, survive and reproduce. In the case of humans, DNA is passed from parents to their offspring. Since all humans come from the same initial population group in Africa, we all carry the genes of our ancestors. By analyzing the genome of the current population, scientists can understand the genome (all genetic information) of our ancestors. This DNA analysis has allowed us to discover that not all of our genome comes from Homo Sapiens. By locating and extracting DNA from a carcass of Homo Neanderthalensis (the Neanderthal) and comparing it to current human DNA, scientists were able to discover that somewhere during ancient human history, humans interbred with the Neanderthals and now up to 4 percent of the human genome comes from another species [??Ovchinnikov, I. et al., Molecular analysis of Neanderthal DNA from the northern Caucasus, Nature, 2000]. Moreover, DNA analysis can reveal exact parts of human history specific to certain regions. For instance, the Denisovans are another species of hominids that interbred with humans, however, their genes can only be found in the people of the Pacific region, indicating that interbreeding between humans and Denisovans happened only in that region [Reich, D. et al., Denisova Admixture and the First Modern Human Dispersals into Southeast Asia and Oceania, American Journal of Human Genetics, 2011].
The second part of the answer lies within DNA’s unique property: the tendency to mutate. A mutation is an alteration of the DNA sequence which may or may not result in changes in the ability of the body to carry out certain functions. Some mutations can be beneficial and result in an evolutionary process, while other mutations are harmful and can cause severe side effects such as cancer and other hereditary diseases. Since mutations can occur down the human family tree and not necessarily in its first ancestors, it allows us to further investigate human history. For example, the Bajau people in Indonesia have a unique DNA mutation that results in enlarged spleens, which are highly beneficial in their fishing lifestyles [Ilardo, M. A. et al, Physiological and Genetic Adaptations to Diving in Sea Nomads, Cell, 2018]. Sequencing DNA allows us to identify all types of mutations and provides crucial information to scientists about the history of particular communities and is especially useful in the field of ecological genetics and anthropology.
The third part of the answer lies in evolutionary science. According to Charles Darwin’s theories, positive selection is the process during which new genetic variations sweep a population. This tendency and the complexity of the evolutionary process can be accessed by taking a look at ancient DNA, or aDNA, (DNA that is derived from ancient specimens). Access to aDNA has allowed scientists to search for specific genes that, through the process of positive selection, allowed modern humans to evolve and outcompete other archaic hominids. Specifically, they helped scientists to comprehend the selective genetic processes that resulted in genetic adaptations by humans to their environments. For example, aDNA allowed scientists to identify specific genes that resulted in the positive selection of taller individuals in the Iberian Neolithic population and the positive selection of shorter individuals in the Steppe Neolithic population [Mathieson, I. et al., Genome-wide patterns of selection in 230 ancient Eurasians, Nature, 2015]. Generally speaking, DNA allowws to identify what mutations superseded others and allowed humans to evolve.
To sum it all up, the history of human evolution and civilization for centuries was surrounded by mystery and speculation, but due to one molecule, DNA, we have learned more and more throughout the years and it is only likely that as time goes on humanity will discover even more about its ancient history.
Essays Honourable Mention 1
Ancient DNA Tells Stories About Hominins
Ibrahim Can Sarikaya, Tuzun Arik Biyikli
Cagribey Anadolu Lisesi, Ankara, Turkey
Archaeologists learn about the past by putting together ancient pieces left behind that tell us how prehistoric humans lived. But imagine being able to study their DNA, to understand how different groups of hominins (modern humans and their extinct ancestors) were related to one another, where they came from, or even what diseases they carried? Nearly two decades ago, researchers found ways for isolating little amount of DNA that was preserved in ancient samples such as bones, teeth, hair, and even sediments. 1 Then they sequenced this ancient DNA (aDNA) by using high-throughput sequencing (HTS) techniques. 1,2,3,4 These genomic discoveries have allowed researchers to compare the genomes of modern humans ( Homo sapiens ) and the genomes of ancient relatives (i.e., Neanderthals and Denisovans). 3,4 For example, if the genome of one group differs from a closely related group by a certain percentage, researchers can estimate how long ago the two groups diverge, knowing the average amount of time it takes mutations to accumulate. By analyzing ancient genomes and comparing them with those of modern humans the researchers can make lots of assumptions about the genetic changes, genetic history of past and present human populations, how these groups actually lived, migrated and interacted with each other. 4,5,6,7 So far from the ancient DNA researches, we have learned that Neanderthals and Denisovans are more closely related to each other than to modern humans. These two archaic descendants are indicated to have separated from modern humans ~550 thousand years ago (ka) and afterward they separated from each other ~400 ka. 2,4 aDNA analysis has also shown that modern humans not only met but mated with Neanderthals after the ~550 ka separation. 2,8,9 The Neanderthals and Denisovans were able to interbreed with each other, as shown by an archaic individual (Denisova 11) who had a Neanderthal mother and Denisovan mother. 10 Aside from expanding our grasp of human history, aDNA research over the last decade has also offered new insights into human biology. A key genetic risk factor for severe COVID-19, for example, is inherited from Neanderthals. 11 Researchers are also looking at what benefits DNA from other ancient humans might have on the health of modern humans. In conclusion, our DNA is like a unique but delicate bridge stretching from past to present. What makes an object antique is a story it carries. Ancient DNA contains stories about human history and by analyzing and comparing ancient genomes we can enhance our understanding of human history and human biology.
Essays Honourable Mention 2
How can DNA help us to discover ancient human history?
Wai Isabel, Astrid Edmunds
Wellington College, Crowthorne, United Kingdom
The use of technology around genome mapping, has been crucial to improving our understanding of our own ancestry. Analysis of the similarities and differences in the DNA of different hominin groups has allowed researchers to map out the tangled family tree of humans 1 . Currently there are three groups of hominin’s that have had their DNA sequenced ; Neanderthals , Denisovans and Homo Sapiens 1 . However these specimens are mostly less than 100,000 years old, when looking further into history our knowledge becomes murkier 1 . While it is possible to identify species present over a million years ago, it is near impossible to trace how these species relate to more contemporary hominins 1 . An example can be seen in H.erectus , which first emerged in Africa around 1.9 million years ago, however it remains uncertain how it is related to H.sapiens 1 . DNA degrades over time, and therefore has left blind spots in history 1 . This in turn has social and political implications as DNA degrades faster in warm environments 1 , this leads to there being massive holes in our knowledge in areas such as Africa or South-East Asia – where the temperatures are much higher 1 . Another flaw in the use of DNA to trace human ancestry, is the possible presence of ‘de novo’ genes 2 . It had previously been believed that evolution occurred through the mutations that cause changes to genes 2 . However when evolutionary biologist Helle Tessand Baalsrud was looking at the genomes of the Atlantic Cod (Gadus Morhua ) she found an antifreeze gene – however when looking at the animal’s relatives she could find none that shared this gene 2 . This led her to suggest that some genes could been seemingly ‘built from scratch’ 3 . The genome itself does not solely consist of DNA that encodes genes 2 . It also consists of long stretches of DNA labelled ‘junk DNA’, that at first glance appears to have no function 2 . However it is now believed that this DNA has the capability to form new functional codes for proteins 2 . It is from this that ‘ De Novo’ genes occur 2 . While the theory of ‘de novo’ genes is essential in the journey to trace human ancestry, as it provides an explanation as to why some present genes cannot be found in any relative species, it still has its limitations. The main limitation is found in the technique used to identify whether the gene is ‘de novo’ or not. The most prevalent technique used is an algorithm that searches all relative species for a similar gene to the one in question, if no such gene is found the gene in question is labelled as ‘de novo’ 2 . The problem is that just because no relatives containing that gene have been found – does not necessarily confirm that no such relatives exist 2 . Due to gaps in our knowledge of different species evolution, it visible to suggest that certain genes and species may have been ‘lost’ over time 2 . For examples one study in 2009 (from Origin of Primate Orphan Genes) found only 15 ‘de novo’ genes within the whole primate order 4 , while a study in 2011 suggested that there were 60 ‘de novo’ genes in humans alone 5 . This large range of possibly answers makes it hard for scientists to accurately trace our ancestry as the conclusions made may be dependent on which pieces of data you use. Another new technique used to help unravel the mystery that is the Homo Sapiens family tree is protein analyse. In the 2000s researches learnt that mass spectrometry could be applied to ancient proteins – the technique involves breaking down proteins into their individual peptides and analysing their masses to help depict their chemical makeup 1 . The benefits of using this technique alongside the genome mapping, is that some proteins are able to withstand time better. Furthermore it allows us to go further back in time and reveal insights about the distant past. An example of this can seen through palaeoproteomics specialist Enrico Capellini’s study of the extinct rhinoceros Stephanorhinus 6 . Through protein analysis he was able to identify the pattern of amino-acid substitutions that suggests that the animal was related to the extinct Woolly Rhinoceros (Coelodonta Antiquitatis) , despite the fact that the extracted proteins were over 1.8million years old 6 . The combination of using protein analyse, scientists knowledge of ‘de novo’ genes, and genome
mapping, allows us to better our understanding of ancient humans and to further discover our own human ancestry.
Essays Honourable Mention 3
CHANGE PROCESSES OF HUMANITY
Nehir Deren FIRAT, EBRU OZDOGRU
SARIYER DOGA HIGH SCHOOL, ISTANBUL, Turkey
CHANGE PROCESSES OF HUMANITY DNA is the material that provides hereditary transmission from a living generation [1]. He takes part in anthropological research on the course of man and his life style. The generality of these societies can be determined by people related to their age world, health problems, diets, livelihoods of people in subsistence [2]. Bone, teeth, dried tissue, hair, coprolite and plant are used as Ancient DNA (aDNA) material [3][4]. Ancient DNA (Ancient DNA), the evolutionary and evolutionary adulthood of DNA, which is a sample from the life of the living, to reveal a wide range of research such as anthropology [5]. The history of ancient DNA began with Higuchi et al.’s 1984 isolation and sequencing of DNA from a museum specimen of a zebra species, through molecular cloning. One of the most important of the aDNA studies is the important study of Svante Paabo and his team in 1985 to obtain 380 base pairs of mitochondrial DNA belonging to a Neanderthal species. Polymerase Chain Reaction (PCR) method developed by K. Mullis in the same years showed that it can produce millions of copies of a special DNA sequence in a very short time [6]. These studies are important because they provide answers to some archeological and anthropological questions such as human change process and migration movements, determination of human-specific genetic and microbial diseases, determination of domesticated animal and plant species [7]. With the increase in ancient DNA studies, it has been possible to compare the data of ancient and modern humans. The information obtained also provides different information about the genetic information they carry, the social and cultural relations of the society in which that individual lives, with which other communities, their lifestyles, nutritional characteristics and adaptation to their environment [8]. Another important study that will shed light on the change process of modern humans is the aDNA data obtained from the finger bone and tooth remains in Denisova Cave in the Altai Mountains. A whole mitochondrial DNA (mtDNA) sequence from the finger bone fossil unearthed in 2008 was analyzed and compared with the mtDNA sequence of 54 modern humans, 6 Neanderthals, and a 30,000-year-old modern human. It has been stated that the Denisova genome contains a different mtDNA sequence from the others. While the Neanderthal mtDNA genome differed from Homo sapiens in an average of 202 nucleotides, the Denisova genome differed in an average of 385 nucleotides. With the mtDNA sequence obtained from this finger bone, it is stated that Denisova human is a different species. Again, studies have shown that the Denisova species are anatomically similar to Neanderthals and modern humans, and that these three species share a common ancestor about 1 million years ago [9][10].
Essays Honourable Mention 4
How DNA can help us to Discover Ancient Human History
Eoghan Strain
Scoil Mhuire Buncrana, Co. Donegal, Ireland
DNA (deoxyribonucleic acid) is the material we inherit from our parents. Most DNA is found in the nucleus of cells however, a small amount is also present in the mitochondria (mtDNA). The information stored in DNA is encoded by four chemical bases: adenine, guanine, cytosine and thymine. The sequence of these bases determines the genetic information in the DNA. [1]
A gene is the basic functional and physical unit of heredity, in essence, they are the blueprints of our bodies and determine each of our physical characteristics. Genes are composed of DNA. Each parent passes down a copy of each gene to their offspring, resulting in us having two copies of each gene. Most genes are the same in everyone however, less than one percent are different. This is called our genetic variation as these small differences determine our unique physical characteristics, for example, our hair and eye colours. [2]
Since our DNA is passed from one generation to the next, our DNA provides a unique insight into our past ancestors. Using mathematics, statistics and computers, scientists can uncover this information and reconstruct DNA family trees by examining the small differences in our DNA. How often we share an ancestor at different times in DNA family trees can tell us how close our ancestors lived together, how they moved about, and how many other humans were alive during that period. Typically, the further back two people share an ancestor, the more genetic differences there are between them. [3]
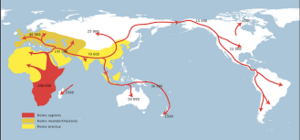
DNA examination has helped us to map the evolution of our ancestors and to form the image below depicting human migration. However, it was only recently discovered through DNA examination that modern humans did not push neanderthals out of Europe, but rather lived alongside them and interbred. [4]
Discoveries of our past are being made constantly, completely transforming our previous understanding of ancient human history. One such discovery is the revelation of an entirely new human species, Desinovans, in 2010. Although the discovery was accidental, these were the first human species to be discovered based on DNA alone. The discovery was made when a German geneticist was examining mtDNA from, whom he thought, a neanderthal. It was only through DNA that he could discover the new species. The later discovery of a fossil with DNA from both Neanderthals and Denisovans proved that these two species interbred with each other. [5]
Recent advances in DNA examination have enabled scientists to examine DNA and make discoveries much more easily. What was impossible just several years ago, is now possible. One such advancement is the invention of polymerase chain reaction (PCR), which has proved instrumental in examining ancient DNA. Since the amount of DNA found in fossils is so small it is very difficult to examine. PCR makes a large number of copies of the gene so it is possible to examine. [6]
Another advancement lies in epigenetics. Because most of the DNA of modern humans is very similar to ancient humans, researchers believe that most of our differences do not come from changes in the DNA itself but changes in DNA regulation (if a gene is turned ‘on’ or ‘off’). [7] In 2014, scientists developed a way to reconstruct epigenetic patterns (DNA methylation), which enables us to understand which genes are expressed and which are silenced. This opens up a range of new possibilities, allowing us to understand how ancient humans differ from modern humans including how they might have looked and sounded, and, in theory, enables us to recreate the environment in which ancient humans lived. [8]
Unfortunately, DNA examination, like everything, has its limits. DNA degrades over time and how well it is preserved is determined by many factors, such as exposure to heat and sunlight. The half-life of DNA is estimated to be around 521 years, and so DNA would not be readable after 1.5 million years in ideal conditions. [9] Although mtDNA is preserved longer than nuclear DNA, it has its limits in that it only provides matrilineal information since it is only inherited from the mother. [10]
Undoubtedly, DNA provides an incredible insight into our ancient history. It has proved fundamental in learning about our ancestors, how they lived and moved about. Despite current limits in DNA examination, recent advancements and trends show that the future of DNA examination looks extremely promising. New technologies continue to open up a range of possibilities for the future.
Essays Honourable Mention 5
How can DNA help us to discover ancient human history?
Anouschka Hartley, Astrid Edmunds
Wellington College, Crowthorne, United Kingdom
Scientists have been aligning DNA sequences since the mid-seventies and from these, they have been able to deduce the lineages of human ancestry. This essay will attempt to evaluate how DNA has helped us understand ancient human historyFirstly, by ‘ancient’ this means history from about 200,000 ago 1 . Researchers are able to look at the similarities and differences of DNA in different hominin groups in order to untangle family trees and to look at our human history of Neanderthals and Denisovans. For example, bones found in a Denisovan cave in Siberia revealed that hominins lived there about 400,000 years ago. A hominin is a primate member of a taxonomic tribe which comprises of species related to humans 2 . The DNA of the bones uncovered that they had come from an unrecognised human lineage that shared the “ice age Eurasia with Neanderthals and modern humans” 3 . Mitochondrial DNA (mtDNA) is very useful in discovering ancient human history as it is inherited solely from your mother, meaning there is less genetic variation. Therefore, it is beneficial when tracking genetic changes over long periods of time as it is more reliable, hence when researchers are studying fossils, they will examine mtDNA rather than nuclear DNA. For example, mtDNA was used to track how humans have expanded out of Africa and it was used to prove that lineages of Neanderthals came out of Africa 4 .There are issues with using DNA to discover ancient human history as it is prone to instability. When comparing DNA with proteins, it degrades faster in warmer environments and so researchers will have missed out DNA of fossils from regions with warmer climates, such as Africa and Asia. As a result, entire sections of human history would have been missed out due to DNA not being durable enough to withstand certain conditions. However, as proteins are much sturdier, due to hydrogen bonds in their tertiary structure, they can be used to fill these gaps by using Zooarchaeology by mass spectrometry. This technique is used to study modern proteins and involves breaking proteins down into short chains of amino acids, to then examine the masses of each chain to determine their chemical make-up. This method has been used to filter through hundreds of bones and researchers can then analyse collagen. They can do this due to the difference in mass of collagen in different species, producing a distinctive fingerprint which can be used by researchers to identify the source of the bones. Therefore, when researchers are unable to recover DNA from certain specimens that are too old or from certain climates that do not favour DNA preservation, proteins can be used as an alternative in discovering ancient human history as they are more stable and could reveal insights into iconic human ancestors 5 .Another issue with using DNA is researchers are unable to uncover the ancient human history of behaviour. Researchers can only use DNA to entice the basic compositions of us, not the history of our behaviour, however artefacts such as cave paintings could help researchers to. For example, a cave painting in Lascaux (see figure 1), estimated to be around 15,000 years old, reveals how humans used to hunt using spears, whereas nowadays we hunt using guns:
Figure 1 6 DNA would only have been able to uncover that humans were alive in that time, however artefacts are able to expand our knowledge further about the history of behaviour, indicating similarities in terms of obtaining food from animals.There are many applications of using DNA for the future. For example, CRISPR can be used to edit a genetic variation, meaning future offspring would not inherit that issue 7 . CRISPR could also be used to kill antibiotic-resistant bacteria in the gut. It has been tested on mice and hopefully it could be transferred and applied to humans as well 8 . However, there are many ethical issues with gene editing, such as the new-born child did not give consent to have their genes edited and changed.In conclusion, DNA is useful in helping researchers analyse the ancient ancestors of humans, discovering what we have evolved from and has many applications for the future. However, the use of proteins may be necessary to fill the gaps in human history where DNA has been destroyed due to the climate.
Essays Honourable Mention 6
How can DNA help us to discover ancient human history?
Flavia Maria Galeazzi, Andrea Bernacchia
ISS Savoia Banincasa, Ancona, Italy
Since 1985, when the DNA of a 2,400-year-old Egyptian child mummy was cloned and sequenced1, the study of human ancient DNA (aDNA) has become of prominent use to identify hominin groups2. Ancient DNA is a versatile tool that provides evidence that directly relates to the organic material, useful for paleontological and historical analysis.Novel technological advancements have greatly simplified and improved sequencing procedures. Nowadays, techniques like Next Generation Sequencing (NGS) provide cost-efficient access to larger genome regions and minimize the contamination and damage impact by using efficient target-enrichment techniques. NGS allowed the analysis of novel bone types and the enhancement of statistical analysis methods2.Genetic data defined answers to questions archeologists debated over for decades, allowing us to look at bigger events rather than focusing on single artifacts. Moreover, genetic methods have been preferred to traditional ones. This aspect further entails an enhancement of the competition for samples and a reduction of evidence for osteological analysis. However, as proven by the Bell-Beaker case3, considering biological evidence superior might risk reducing the complexity of human actions to a genetic map, while archeological evidence may not always be consistent with ethnic presence. In recent years, there has been an increasing number of cross-field publications, thus proving how aDNA is driving science towards a more cooperative approach.This has brought about advancements in investigating extinct hominid genetic introgressions, language, and human pathogen evolution.Neanderthal ancestry was found in gene-regulating sequences and has been linked to depression, skin phenotypes, and blood4. aDNA is utilized to determine complex-trait heritage in modern humans, particularly standing height, skin, and eye pigmentation5.Particularly, the Val92Met loss-of-function mutation in the melanocortin 1 receptor (MRC1) caused Neanderthals to evolve particular photoaging and skin-color phenotypes. However, since no modern human possesses this exact mutation, Homo Sapiens and Neanderthals possibly evolved the phenotype independently as a response to sunlight intensity6. Blood type inheritance is key in determining Neanderthal ancestry. Neanderthals and Denisovans possess polymorphic ABO-related alleles, also found in modern humans. The evolutionary advantage was probably the prevention of viral gut infection. The presence of RHD and RHCE alleles correlates with a predisposition towards fetal hemolytic disease, thus hypothesizing a reason for Neanderthal extinction7.Evidence of immune system haplotype introgression and positive selection has been linked to HLA haplotypes and Toll-like receptors8,9. Particularly, TLR1, TLR6, and TLR10 receptors, encoded in the same cluster, are responsible for first-line defense against pathogens and for the activation of innate immune response9.The FOXP2 gene has been linked to speech and language10. Both Neanderthals and modern humans seem to carry the same gene mutation, thus suggesting that the selective sweep happened before the two lines diverged. By triangulating the aDNA data with Bayesian linguistic analysis, it was inferred that genetically-related ethnic groups shared a common language11. Moreover, the analysis of DNA encoding throat structure provides evidence for linguistic evolution12.NOVA-1 gene variants and their neurodevelopmental implications are being studied on cortical organoids “to explore other genetic changes that underlie the phenotypic traits separating our species from extinct relatives”13.Pathogens are considered a potent driver for adaptation. In the Neolithic period, domestication provided new opportunities for zoonotic pathogenic transfer. Ancient DNA has proved key in dating the origin of the M. tuberculosis complex (MTBC), which appeared to be correlated with farming and agriculture development in the Neolithic period14. aDNA helped reveal how Y. Pestis, an environmental bacterium, and opportunistic gastroenteric pathogen, evolved into an extremely contagious form by acquiring just a few virulent factors and seems to be responsible for the Neolithic decline15. aDNA has revolutionized our understanding of the historical emergence and spread of infectious diseases. Advancements in disease modeling and genome sequencing can also give insight into migratory patterns and single-case archeological analysis.However, comparing modern with ancient ethnic groups may prove problematic16. Attributing human ethical status to a specimen is another question17. For example, a not-identified specimen who was deemed of alien origin was later identified as a girl18. aDNA evidence is contributing to determining a better definition of the human species in genetic and evolutionary terms. Conducting research near indigenous communities may undermine deeply held beliefs, encourage disagreements among groups, and complicate land property issues19. When aDNA is studied, the bone fragment is often destroyed. Nowadays, scientists are increasingly prone to connect with local stakeholders and assure the establishment of informed consent before starting scientific investigations16,17.What aDNA can tell us greatly supersedes its potential drawbacks, and its revolutionary impact on many fields has provided unprecedented opportunities for investigating human evolution in different ways.
Essays Honourable Mention 7
How can DNA help us to discover ancient human history?
Charlie Gould, Astrid Edmunds
Wellington College, Crowthorne, United Kingdom
Deoxyribonucleic acid, DNA, is found in nearly every cell in a human’s body and contains nearly 3 billion bases that determines our unique genetic code (1) . The genetic code is like a manual, providing instructions on producing amino acids to form proteins. DNA was extracted from the bones of the extinct moa bird in a study conducted in New Zealand, found that DNA has a half life of 521 years, meaning that 75% of genetic information is lost after 1000 years, and all genetic information is lost after 6.8 million years ( 2 ) . Within this timeframe, scientists are able to conduct studies on the DNA of fossils, and this is vital in discovering ancient human history. DNA can be used to locate the origins of human ancestors too. The DNA of people living in the British Isles was studied by scientists, and they found that there were differences in the DNA of groups of people who only lived short distances apart. They were able, for example, to find the border between Cornwall and Devon, because the DNA of someone who’s grandparents were born in Cornwall was different to the DNA of someone whos’ grandparents were born in Devon. This created areas of relative isolation, as the ancestors of people in Devon would have only had children with other people from Devon, and not crossed into Cornwall. There are other areas of relative isolation due to geographical isolation such as the Orkney islands, or due to certain historical events, such as settlements of Britons and Saxons. ( 3 ) The DNA from areas of relative isolation is pivotal in determining movement of ancient humans. Geographical features, such as vast waterways or mountain ranges, can prevent the migration of people, and thus can create areas of relative isolation. This can, of course, increase extinction risk within that group, because genetic diversity decreases, and inbreeding can lead to deadly mutations within communities. But, there are many examples of human groups migrating away from isolated areas, often across large oceans or vast mountain ranges, and this can be seen in the DNA of living people today. An example of this is Iberia, where it is known that during the time of Muslim rule around 1000 CE, there was a movement of people from North Africa into Iberia. When DNA of Spanish people today was compared with people in other countries around Europe and North Africa, it was clear that those North African migrants would have had children with the locals in Iberia. ( 3 ) The science behind ancient DNA is still reasonably new. In 2010, the first fully sequenced ancient human genome (‘ancient DNA refers to the study of DNA extracted from specimens that died decades, hundreds or sometimes thousands of years ago’ ( 4 ) )was produced. Research was limited to skeletons from cold climates, as DNA is more likely to preserve there. However, a recent breakthrough has been highly important in expanding the range of skeletons that can be studied. The bony casing around the inner ear, known as the petrous, is a very rich source of ancient DNA, even in skeletons that were from hot climates and have been badly preserved. This has meant that there has been a massive increase in the pace of studies, “with thousands of individuals sequenced in 2018”. ( 5 ) Although DNA can be very useful in discovering ancient human history, proteins within fossils may be more useful. A study conducted on a hominin’s teeth found that its “chemical signature was a single amino acid variant that isn’t present in the collagen of modern humans or Neanderthals”. Instead, the jawbone of the hominin belonged to a member of a group called Denisovans. This was the first time that only proteins had been used to identify an ancient hominin. The study put forward an argument that proteins may be more useful for identifying ancient human history, as proteins remain in fossils for much longer than DNA does, because in a particular study, it was found that ‘the proteins in (her) 3.8 million year old eggshell had bound to the surface of the mineral crystals in the shell, essentially freezing them in place.’ (6) This essentially means that proteins can bind to
minerals which then protects them from degradation, and thus supports the claim that proteins can be used when all DNA has degraded. This new research is allowing a whole new area of prehistory to be explored by scientists. ( 6 )
Essays Honourable Mention 8
IS ANCIENT HISTORY ENCODED IN OUR DNA?
Melda Parlakisik
The Koc School, Istanbul, Turkey
Deoxyribose nucleic acid (DNA) is a nucleic acid that contains genetic information consisting of units called nucleotides found in the structure of organisms [1,2]. Due to the information it contains, DNA enables the creation of cell components such as proteins and RNA. However, DNA is also responsible for where and how genetic information is used [2]. Is ancient history encoded in our DNA? Ancient DNA studies were defined as DNA extraction performed on damaged biological samples and not kept under special conditions for DNA extraction [3]. The first DNA studies in literature were the studies on the quagga, which is considered to be the ancestor of the donkey species, by Higuchi et al. (1984), and Paabo et al. (1985) studied on an Egyptian mummy [4,5]. With the technological developments and increasing media attention since the discovery of DNA in the 1980s, researchers from many different scientific disciplines started to focus on DNA to learn about history. Ancient DNA findings have been a light in illuminating the dark ages of the past. Neanderthals, classified as a human species and called “Homo neanderthalensis”, lived as hunters and gatherers between 400,000-30,000 years ago. The Neanderthal genome project shows that modern humans began to diverge from the descendants of Neanderthals about half a million years ago [6]. It is known that modern humans (modern Homo sapiens) emerged in Africa 200,000 years ago. Despite this long period of time, some studies continue to establish genetic similarities between Neanderthals and modern humans. It has been reported that 1-4% of the DNA of people living outside Africa belongs to Neanderthals [7]. Genetic similarity also helps to understand history better. It has been reported that DNA traces of Denisova people, who lived in Southern Siberia 40,000 years ago, known as relatives of Neanderthals and modern humans, are found in Malaysian residents thousands of kilometers from Siberia today. This information indicates that Denisovans once lived in Asia [7,8]. When Abi-Rached et al (2011) compared the human leukocyte antigen (HLA) gene, which is an immune gene of Neanderthals, Denisovans and modern humans, they concluded that this gene is more common in modern humans living in parts of Europe and Asia than in Africans. However, it was understood that Neanderthals and Denisovans also carried the same gene. This result supports the idea that both species may have once lived in Asia. Also, Abi-Rached et al. (2011) showed that the genes that enable humans to fight diseases today came from Neanderthals and Denisovans [9]. On the other hand, Hawsk (2017) reported that Neanderthals and Denisovans did not have DNA forms that would enable them to defend against diseases such as measles, like modern humans. Researchers explain this by the fact that the people of that period lived as hunters and gatherers but populations did not assemble. Accordingly, mass epidemics do not occur frequently, and there was no need to develop immune genes [10]. Analyzing the genetic information of pre-BC humans in the British region, Brace et al. (2019) determined that there was little biological continuity between groups living in the Mesolithic and Neolithic periods. This situation led the researchers to think that there was a great population change at that time. Researchers argued that this change was achieved through migration, and migration caused people of the region to start farming [11]. Similarly, Olalde et al. (2018) argued that the DNA signature of the Neolithic population in Britain decreased, and this was due to the influx of immigrants [12]. Another study states that the factor that triggered the start of farming in Europe in the Neolithic period was the migration from Anatolia [13]. The study of Allentoft et al. (2015) showed that there was a harmony between migrations and cultural changes in the Bronze Age [14]. From this, it can be said that DNA research has an important role in understanding the migration periods and the reasons for the transition to agriculture. In conclusion, it is clear that DNA is an effective method for discovering ancient human history. DNA provides excessive information such as where ancient people lived, when they migrated, how these migrations affected other species and how these species lived. In addition, DNA also helps explain the connection of today’s people with their past, their ancestors, and the evolutionary process. Considering that the study of ancient DNA has a history of approximately 30 years, it is in fact, a new field. With the development of technology over time, DNA will have a lot to teach and explain about history.
Essays Honourable Mention 9
We can now sequence the genome of all life forms, from viruses to humans. What could be the point of this?
Simona Zelionkaite, Egle Tauraite
Vilnius Lyceum, Vilnius, Lithuania
I propose to sequence the African genome because of its impressive genetic diversity. Establishing a detailed understanding of the heritable variation in the human genome is one of the key challenges of the post-genome period. Progress in human genome research is currently being observed, but while the importance of genetic data is increasing, the problem is not the availability of sequencing but the lack of diversity in genetic information. Since modern people originated in Africa and have adjusted to assorted conditions [1], African populations have substantially higher levels of genetic and phenotypic diversity. Subsequently, it is fundamental to study genomic diversity of African ethnic groups in order to understand human evolutionary history and how this prompts differential disease risk in all people. Africa is a diverse region in terms of genetics, culture, semantics and phenotypic characteristics. There are over 2000 specific ethnolinguistic groups in Africa, communicating in dialects that make up about almost 33% of the world’s languages. The genetic diversity of Africa is influenced by a variety of factors [2]. To begin, Africans live in a wide range of habitats ranging from deserts and tropical rainforests to savannahs and wetlands. Secondly, different exposures to infectious diseases is also a major factor which determines genetic diversity. And lastly, Africans have diversified subsistence and diet patterns that include pastoralism or hunting-gathering. To summarize, Africa is so diverse because of its size – Africa is about the size of the land masses of China, the United States, India, Japan, and most of Europe combined. It’s a continent consisting of 54 countries which stand out both culturally and geographically. Taking this into account, it can be said that broadening the genetic data with African genomic information could utterly transform the world. There are two potential scenarios – realizing that the African genetic data could possibly change our whole perception of disease prevention and treatment or ignoring this fact and facing stigma. Nowadays most of the ongoing genetic studies rely mostly on European genetic information. Current lack of genetic diversity is one of the main reasons why scientists fail to incorporate genomic research into clinical practice. The purpose of all medicine can be described in three words: diagnose, treat, prevent. Medicine, with all the necessary data and using certain existing genetic markers, could adapt the diagnosis and treatment of the disease according to a person’s unique genetic model. For the time being, however, scientists are conducting research largely based on data from white European populations, distinguishing Africans that have a wide genetic variety. Such a lack of diversity in genomics research is thought to undermine our scientific understanding of the genetic basis of disease in all populations and increase health care bias. Take, for example, a single mutation named ?F508 in CFTR gene which accounts for 70% cystic fibrosis (CF) cases in the world [3,4]. In this case, the disease is caused by a specific variant of one gene, which in one population should be as disease-causing as in other populations, but this does not always correspond to real cases of the disease. This mutation is responsible for more than 70% of CF cases in Europeans, but only for 29% of cases in the African diaspora. Thus, even single gene mutations in
different populations have different consequences for the phenotype, and the lack of diversity of genetic data potentially affects many non-Europeans without adequate diagnostic and treatment possibilities. Limiting oneself to the genome information of Europeans greatly narrows the horizons of scientists and prevents other populations from receiving full-fledged treatment. It is clear that different genetic variations in different populations can affect both disease risk and treatment efficacy. Unfortunately, most research in this area is still concentrated on populations of European ancestry, as approximately 80% of participants in genomics research are of European descent [5], and the results obtained are very limited and not necessarily meaningfully transferable to other populations such as Africans. This trend is reflected in the poor diagnosis of diseases in individuals who are underrepresented in such studies. By placing utmost importance on diversity in genetic and genomic research, we would improve our ability to have a broader understanding of genetic disease architecture, which would ultimately increase the accuracy of medical care.
References:[1] Schmid, R., 2009. Africans have world’s greatest genetic variation. [online] nbcnews.com. Available at: <https://www.nbcnews.com/id/wbna30502963> [Accessed 24 April 2021].[2] Campbell, M. and Tishkoff, S., 2008. African Genetic Diversity: Implications for Human Demographic History, Modern Human Origins, and Complex Disease Mapping. [online] ncbi.nlm.nih.gov. Available at: <https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2953791/> [Accessed 24 April 2021]. [3] Lukacs, G. and Verkman, A., 2011. CFTR: folding, misfolding and correcting the ?F508 conformational defect. [online] ncbi.nlm.nih.gov. Available at: <https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3643519/> [Accessed 24 April 2021]. [4] Alfonso-Sánchez, M., Pérez-Miranda, A., García-Obregón, S. and Peña, J., 2011. An evolutionary approach to the high frequency of the Delta F508 CFTR mutation in European populations. [online] NIH. Prieinamas: <https://pubmed.ncbi.nlm.nih.gov/20110149/> [Accessed 24 April 2021].[5] Hussein, S., 2020. New genome sequencing sheds light on diversity in Africa. [online] medicalxpress.com. Available at: <https://medicalxpress.com/news/2020-10-major-african-genome-varieties-history.html> [Accessed 24 April 2021].